Validation of the potentials
Contents
Validation of the potentials#
Once we have the fitted potentials, it is necessary to validate them in order to assess their quality with respect to applications.
In this exercise, we use the fitted potentials and perform some basic calculations.
Import the fitted potentials for Li-Al (from earlier excercise)#
The same directory contains a helper.py file which among other things, also contains the necessary specifications of each of the potentials that we will use today. Individual potentials are descrbed in the LAMMPS format as:
pot_eam = pd.DataFrame({
'Name': ['LiAl_eam'],
'Filename': [["../potentials/AlLi.eam.fs")]],
'Model': ["EAM"],
'Species': [['Li', 'Al']],
'Config': [['pair_style eam/fs\n', 'pair_coeff * * AlLi.eam.fs Li Al\n']]
})
A list of such DataFrames describing the potentials is saved in a list called potentials_list. We import the list as:
from helper import potentials_list
# potentials_list = [potentials_list[0],potentials_list[1]]
# display the first element in the list
# which is an EAM potential
potentials_list[2]
| Name | Filename | Model | Species | Config | |
|---|---|---|---|---|---|
| 0 | LiAl_yace | [/home/jovyan/workshop_preparation/potentials/... | ACE | [Al, Li] | [pair_style pace\n, pair_coeff * * AlLi-6gen-1... |
Import other important modules#
import numpy as np
import matplotlib.pylab as plt
import seaborn as sns
import pandas as pd
import time
from helper import get_clean_project_name
from pyiron_atomistics import Project
from pyiron import pyiron_to_ase
import pyiron_gpl
# save start time to record runtime of the notebook
time_start = time.time()
time_start
1654697715.1585205
Create a new project to perform validation calculations#
It is useful to create a new project directory for every kind of calculation. Pyiron will automatically create subdirectories for each potential and property we calculate.
pr = Project("validation_LiAl")
# remove earlier jobs
# pr.remove_jobs(silently=True, recursive=True)
Define the important pases to consider for validation#
We construct a python dictionary struct_dict which contains a description of all the important phases that we want to consider for this exercise. The descriptions given in the dictionary will be later used by Pyiron to generate or read the structural configurations for the respective phases.
For unary phases, we provide an initial guess for the lattice parameter and use pyiron to generate the structural prototype.
For binary phases, we provide a phase name and an additional dictionary fl_dict which maps the phase name to a .cif file saved in a subdirectory. Pyiron will use this information to read the respective configurations from the file.
struct_dict = dict()
# structures to be generated automatically
struct_dict["Al"] = dict()
struct_dict["Al"]["s_murn"] = ["fcc","bcc"]
struct_dict["Al"]["a"] = 4.04
struct_dict["Li"] = dict()
struct_dict["Li"]["s_murn"] = ["bcc","fcc"]
struct_dict["Li"]["a"] = 3.5
# structures to be read from file
struct_dict["Li2Al2"] = dict()
struct_dict["Li2Al2"]["s_murn"] = ["Li2Al2_cubic"]
struct_dict["LiAl3"] = dict()
struct_dict["LiAl3"]["s_murn"] = ["LiAl3_tetragonal"]
struct_dict["LiAl3"] = dict()
struct_dict["LiAl3"]["s_murn"] = ["LiAl3_cubic"]
struct_dict["Li9Al4"] = dict()
struct_dict["Li9Al4"]["s_murn"] = ["Li9Al4_monoclinic"]
struct_dict["Li3Al2"] = dict()
struct_dict["Li3Al2"]["s_murn"] = ["Li3Al2_trigonal"]
struct_dict["Li4Al4"] = dict()
struct_dict["Li4Al4"]["s_murn"] = ["Li4Al4_cubic"]
struct_dict
{'Al': {'s_murn': ['fcc', 'bcc'], 'a': 4.04},
'Li': {'s_murn': ['bcc', 'fcc'], 'a': 3.5},
'Li2Al2': {'s_murn': ['Li2Al2_cubic']},
'LiAl3': {'s_murn': ['LiAl3_cubic']},
'Li9Al4': {'s_murn': ['Li9Al4_monoclinic']},
'Li3Al2': {'s_murn': ['Li3Al2_trigonal']},
'Li4Al4': {'s_murn': ['Li4Al4_cubic']}}
a dictionary is described to map the binary phases to their file locations
fl_dict = {"Li2Al2_cubic": "mp_structures/LiAl_mp-1067_primitive.cif",
"LiAl3_tetragonal":"mp_structures/LiAl3_mp-975906_primitive.cif",
"LiAl3_cubic":"mp_structures/LiAl3_mp-10890_primitive.cif",
"Li9Al4_monoclinic":"mp_structures/Li9Al4_mp-568404_primitive.cif",
"Li3Al2_trigonal":"mp_structures/Al2Li3-6021.cif",
"Li4Al4_cubic":"mp_structures/LiAl_mp-1079240_primitive.cif"}
Visualize the strucs#
Once the structures are defined in the pyiron format, we can view their atomic coordinates and cell vectors using struc.plot3d()
# Option 1: use `ase.build.bulk` functionality in pyiron
struc = pr.create_ase_bulk("Al", "fcc", a=4.04,cubic=True)
# struc.plot3d()
# Option 2: Read from a file
struc = pr.create.structure.ase.read(fl_dict["Li4Al4_cubic"])
# struc.plot3d()
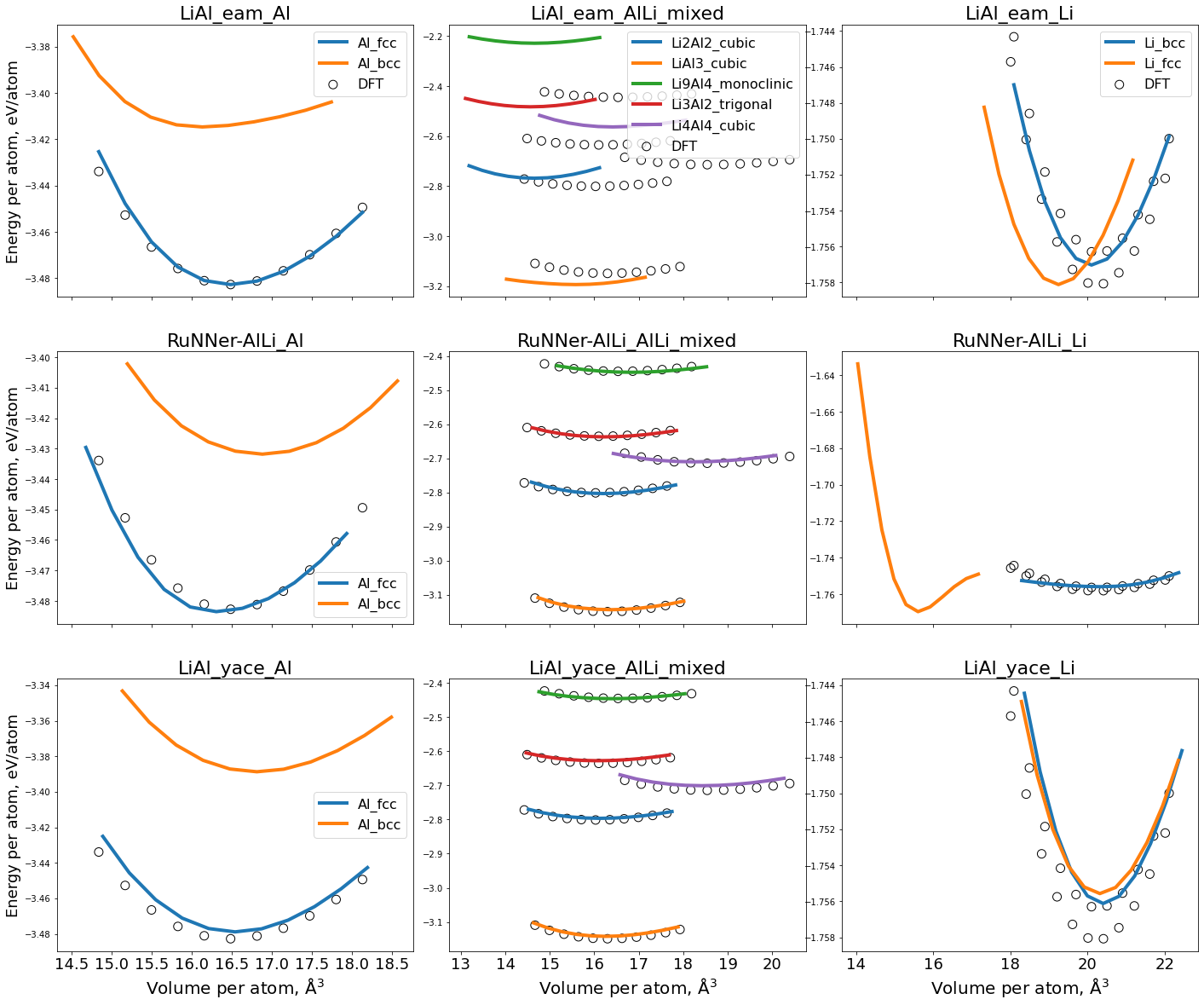
(a) Ground state: E-V curves#
Using a series of nested for loops, we calculate the murnaghan EV-curves using all three potentials for all the defined structures.
We loop over:
All the potentials defined in
potentials_listand name the project according to the potentialAll the chemical formulae defined in the keys of
struct_dictAll phases defined for a given chemical formula
Within the loops, the first step is to get the structure basis on which we will perform the calculations.
For unary phases, we use the pyiron function
pr_pot.create_ase_bulk(compound, crys_structure, a=compound_dict["a"])For binary structures, we read the basis using
pr.create.structure.ase.read(fl_path)with thefl_pathgiven byfl_dictdefined earlier.
Once the structure and potential is defined as part of the pr_job, we run two calculations:
job_relaxto relax the structure to the ground statemurn_jobto calculate the energies in a small volume range around the equilibrium
As the calculations are being performed, the status(s) of each calculation is printed. If a job is already calculated, the calculations are not re-run but rather re-read from the saved data.
for pot in potentials_list:
with pr.open(get_clean_project_name(pot)) as pr_pot:
print(pr_pot)
for compound, compound_dict in struct_dict.items():
for crys_structure in compound_dict["s_murn"]:
# Relax structure
if crys_structure in ["fcc","bcc"]:
basis = pr_pot.create_ase_bulk(compound, crys_structure, a=compound_dict["a"])
else:
basis = pr_pot.create.structure.ase.read(fl_dict[crys_structure])
job_relax = pr_pot.create_job(pr_pot.job_type.Lammps, f"{compound}_{crys_structure}_relax", delete_existing_job=True)
job_relax.structure = basis
job_relax.potential = pot
job_relax.calc_minimize(pressure=0)
job_relax.run()
# Murnaghan
job_ref = pr_pot.create_job(pr_pot.job_type.Lammps, f"ref_job_{compound}_{crys_structure}")
job_ref.structure = job_relax.get_structure(-1)
job_ref.potential = pot
job_ref.calc_minimize()
murn_job = job_ref.create_job(pr_pot.job_type.Murnaghan, f"murn_job_{compound}_{crys_structure}")
murn_job.input["vol_range"] = 0.1
murn_job.run()
/home/jovyan/workshop_preparation/validation/validation_LiAl/LiAl_eam/
The job Al_fcc_relax was saved and received the ID: 1752
2022-06-08 14:15:16,704 - pyiron_log - WARNING - The job murn_job_Al_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Al_bcc_relax was saved and received the ID: 1753
2022-06-08 14:15:18,339 - pyiron_log - WARNING - The job murn_job_Al_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li_bcc_relax was saved and received the ID: 1754
2022-06-08 14:15:19,051 - pyiron_log - WARNING - The job murn_job_Li_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li_fcc_relax was saved and received the ID: 1755
2022-06-08 14:15:20,323 - pyiron_log - WARNING - The job murn_job_Li_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li2Al2_Li2Al2_cubic_relax was saved and received the ID: 1756
2022-06-08 14:15:22,012 - pyiron_log - WARNING - The job murn_job_Li2Al2_Li2Al2_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job LiAl3_LiAl3_cubic_relax was saved and received the ID: 1757
2022-06-08 14:15:23,393 - pyiron_log - WARNING - The job murn_job_LiAl3_LiAl3_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li9Al4_Li9Al4_monoclinic_relax was saved and received the ID: 1758
2022-06-08 14:15:24,515 - pyiron_log - WARNING - The job murn_job_Li9Al4_Li9Al4_monoclinic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li3Al2_Li3Al2_trigonal_relax was saved and received the ID: 1759
2022-06-08 14:15:25,720 - pyiron_log - WARNING - The job murn_job_Li3Al2_Li3Al2_trigonal is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li4Al4_Li4Al4_cubic_relax was saved and received the ID: 1760
2022-06-08 14:15:27,012 - pyiron_log - WARNING - The job murn_job_Li4Al4_Li4Al4_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
/home/jovyan/workshop_preparation/validation/validation_LiAl/RuNNer-AlLi/
The job Al_fcc_relax was saved and received the ID: 1761
2022-06-08 14:15:27,799 - pyiron_log - WARNING - The job murn_job_Al_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Al_bcc_relax was saved and received the ID: 1762
2022-06-08 14:15:28,779 - pyiron_log - WARNING - The job murn_job_Al_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li_bcc_relax was saved and received the ID: 1763
2022-06-08 14:15:29,535 - pyiron_log - WARNING - The job murn_job_Li_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li_fcc_relax was saved and received the ID: 1764
2022-06-08 14:15:30,637 - pyiron_log - WARNING - The job murn_job_Li_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li2Al2_Li2Al2_cubic_relax was saved and received the ID: 1765
2022-06-08 14:15:32,209 - pyiron_log - WARNING - The job murn_job_Li2Al2_Li2Al2_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job LiAl3_LiAl3_cubic_relax was saved and received the ID: 1766
2022-06-08 14:15:33,439 - pyiron_log - WARNING - The job murn_job_LiAl3_LiAl3_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li9Al4_Li9Al4_monoclinic_relax was saved and received the ID: 1767
2022-06-08 14:15:34,543 - pyiron_log - WARNING - The job murn_job_Li9Al4_Li9Al4_monoclinic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li3Al2_Li3Al2_trigonal_relax was saved and received the ID: 1768
2022-06-08 14:15:35,739 - pyiron_log - WARNING - The job murn_job_Li3Al2_Li3Al2_trigonal is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li4Al4_Li4Al4_cubic_relax was saved and received the ID: 1769
2022-06-08 14:15:36,881 - pyiron_log - WARNING - The job murn_job_Li4Al4_Li4Al4_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
/home/jovyan/workshop_preparation/validation/validation_LiAl/LiAl_yace/
The job Al_fcc_relax was saved and received the ID: 1770
2022-06-08 14:15:38,178 - pyiron_log - WARNING - The job murn_job_Al_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Al_bcc_relax was saved and received the ID: 1771
2022-06-08 14:15:39,859 - pyiron_log - WARNING - The job murn_job_Al_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li_bcc_relax was saved and received the ID: 1772
2022-06-08 14:15:41,447 - pyiron_log - WARNING - The job murn_job_Li_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li_fcc_relax was saved and received the ID: 1773
2022-06-08 14:15:42,997 - pyiron_log - WARNING - The job murn_job_Li_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li2Al2_Li2Al2_cubic_relax was saved and received the ID: 1774
2022-06-08 14:15:44,100 - pyiron_log - WARNING - The job murn_job_Li2Al2_Li2Al2_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job LiAl3_LiAl3_cubic_relax was saved and received the ID: 1775
2022-06-08 14:15:45,161 - pyiron_log - WARNING - The job murn_job_LiAl3_LiAl3_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li9Al4_Li9Al4_monoclinic_relax was saved and received the ID: 1776
2022-06-08 14:15:46,678 - pyiron_log - WARNING - The job murn_job_Li9Al4_Li9Al4_monoclinic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li3Al2_Li3Al2_trigonal_relax was saved and received the ID: 1777
2022-06-08 14:15:48,152 - pyiron_log - WARNING - The job murn_job_Li3Al2_Li3Al2_trigonal is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
The job Li4Al4_Li4Al4_cubic_relax was saved and received the ID: 1778
2022-06-08 14:15:49,387 - pyiron_log - WARNING - The job murn_job_Li4Al4_Li4Al4_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
One can display the technical details of all submitted jobs using pr.job_table() below.
# pr.job_table()
In order to get read useful results from the completed calculations (eq_energy, eq_volume, etc), it is useful to define the following functions
# Only work with Murnaghan jobs
def get_only_murn(job_table):
return (job_table.hamilton == "Murnaghan") & (job_table.status == "finished")
def get_eq_vol(job_path):
return job_path["output/equilibrium_volume"]
def get_eq_lp(job_path):
return np.linalg.norm(job_path["output/structure/cell/cell"][0]) * np.sqrt(2)
def get_eq_bm(job_path):
return job_path["output/equilibrium_bulk_modulus"]
def get_potential(job_path):
return job_path.project.path.split("/")[-3]
def get_eq_energy(job_path):
return job_path["output/equilibrium_energy"]
def get_n_atoms(job_path):
return len(job_path["output/structure/positions"])
def get_ase_atoms(job_path):
return pyiron_to_ase(job_path.structure).copy()
def get_potential(job_path):
return job_path.project.path.split("/")[-2]
def get_crystal_structure(job_path):
return job_path.job_name.split("_")[-1]
def get_compound(job_path):
return job_path.job_name.split("_")[-2]
Using the functions defined above, one can now define a pd.DataFrame containing all useful results
# Compile data using pyiron tables
table = pr.create_table("table_murn", delete_existing_job=True)
table.convert_to_object = True
table.db_filter_function = get_only_murn
table.add["potential"] = get_potential
table.add["ase_atoms"] = get_ase_atoms
table.add["compound"] = get_compound
table.add["crystal_structure"] = get_crystal_structure
table.add["a"] = get_eq_lp
table.add["eq_vol"] = get_eq_vol
table.add["eq_bm"] = get_eq_bm
table.add["eq_energy"] = get_eq_energy
table.add["n_atoms"] = get_n_atoms
table.run()
data_murn = table.get_dataframe()
data_murn["phase"] = data_murn.compound + "_" + data_murn.crystal_structure
data_murn
The job table_murn was saved and received the ID: 1779
/srv/conda/envs/notebook/lib/python3.8/site-packages/pyiron_base/table/datamining.py:620: PerformanceWarning:
your performance may suffer as PyTables will pickle object types that it cannot
map directly to c-types [inferred_type->mixed,key->block2_values] [items->Index(['potential', 'ase_atoms', 'compound', 'crystal_structure'], dtype='object')]
self.pyiron_table._df.to_hdf(
| job_id | potential | ase_atoms | compound | crystal_structure | a | eq_vol | eq_bm | eq_energy | n_atoms | phase | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1140 | LiAl_eam | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.039967 | 16.495612 | 85.876912 | -3.483097 | 1 | Al_fcc |
| 1 | 1153 | LiAl_eam | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | bcc | 3.898853 | 16.147864 | 48.620841 | -3.415312 | 1 | Al_bcc |
| 2 | 1166 | LiAl_eam | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | bcc | 4.195477 | 20.114514 | 13.690609 | -1.757011 | 1 | Li_bcc |
| 3 | 1179 | LiAl_eam | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | fcc | 4.253841 | 19.241330 | 13.985972 | -1.758107 | 1 | Li_fcc |
| 4 | 1192 | LiAl_eam | (Atom('Li', [4.359978178265943, 2.5172345748814795, 1.7799536377360747], index=0), Atom('Li', [6.53996726740165, 3.775851862320358, 2.669930456604317], index=1), Atom('Al', [-3.964456982410852e-12... | Li2Al2 | cubic | 6.165940 | 58.604895 | 100.347240 | -11.074362 | 4 | Li2Al2_cubic |
| 5 | 1205 | LiAl_eam | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [1.9825515172760235, 1.9825515172760237, 2.427925369776811e-16], index=1), Atom('Al', [1.9825515172760235, 1.2139626848884054e-16, 1.9825515172760... | LiAl3 | cubic | 5.607502 | 62.227580 | 51.472656 | -12.774590 | 4 | LiAl3_cubic |
| 6 | 1218 | LiAl_eam | (Atom('Li', [4.9874611628416465, 1.0099045365192156, 0.8188840806477526], index=0), Atom('Li', [3.1237816780987666, 1.455730745331952, 2.673723152073369], index=1), Atom('Li', [-3.4421956688209843... | Li9Al4 | monoclinic | 13.023701 | 190.504374 | 53.125276 | -28.970054 | 13 | Li9Al4_monoclinic |
| 7 | 1231 | LiAl_eam | (Atom('Al', [2.1548001975659234, 1.244075358781918, 1.861784175000869], index=0), Atom('Al', [-2.154798282819334, 3.732223313213554, 2.6646760238080542], index=1), Atom('Li', [8.560563403365654e-0... | Li3Al2 | trigonal | 6.094693 | 72.810229 | 69.231669 | -12.413856 | 5 | Li3Al2_trigonal |
| 8 | 1244 | LiAl_eam | (Atom('Li', [2.142967147985671, 1.2372426587287435, 7.662120717536293], index=0), Atom('Li', [-8.783761113500244e-10, 2.4744853189563414, 0.5913679335098909], index=1), Atom('Li', [-8.783761113500... | Li4Al4 | cubic | 6.061226 | 131.389799 | 71.221355 | -20.506570 | 8 | Li4Al4_cubic |
| 9 | 1257 | RuNNer-AlLi | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.025259 | 16.355737 | 76.669339 | -3.484016 | 1 | Al_fcc |
| 10 | 1270 | RuNNer-AlLi | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | bcc | 3.958447 | 16.870137 | 51.052272 | -3.432183 | 1 | Al_bcc |
| 11 | 1283 | RuNNer-AlLi | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | bcc | 4.211118 | 20.286595 | 8.517306 | -1.755918 | 1 | Li_bcc |
| 12 | 1296 | RuNNer-AlLi | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | fcc | 3.967043 | 15.678901 | 147.215464 | -1.769260 | 1 | Li_fcc |
| 13 | 1309 | RuNNer-AlLi | (Atom('Li', [4.509081801264686, 2.603319591757272, 1.8408249369278522], index=0), Atom('Li', [6.763622701898693, 3.90497938763465, 2.7612374053913604], index=1), Atom('Al', [-3.844724064520768e-12... | Li2Al2 | cubic | 6.376805 | 64.816143 | 57.934650 | -11.212634 | 4 | Li2Al2_cubic |
| 14 | 1322 | RuNNer-AlLi | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [2.0154153406879987, 2.0154153406879987, 2.46817194592603e-16], index=1), Atom('Al', [2.0154153406879987, 1.234085972963015e-16, 2.015415340687998... | LiAl3 | cubic | 5.700455 | 65.403086 | 59.308440 | -12.574696 | 4 | LiAl3_cubic |
| 15 | 1335 | RuNNer-AlLi | (Atom('Li', [5.206051477294367, 1.0619663179427192, 0.8311820920214751], index=0), Atom('Li', [3.28638171437237, 1.5211864250363467, 2.7226207058417775], index=1), Atom('Li', [-3.6198784902055765,... | Li9Al4 | monoclinic | 13.640614 | 218.932018 | 33.874957 | -31.820765 | 13 | Li9Al4_monoclinic |
| 16 | 1348 | RuNNer-AlLi | (Atom('Al', [2.2338755345732753, 1.289729472183878, 1.9126243306628208], index=0), Atom('Al', [-2.233873547699001, 3.869185551846968, 2.7799443936883206], index=1), Atom('Li', [9.007133262260959e-... | Li3Al2 | trigonal | 6.318351 | 81.143544 | 44.574696 | -13.185198 | 5 | Li3Al2_trigonal |
| 17 | 1361 | RuNNer-AlLi | (Atom('Li', [2.220260976080854, 1.2818682724036983, 7.872085429446316], index=0), Atom('Li', [1.722758777253687e-10, 2.5637365444716322, 0.6790950189344616], index=1), Atom('Li', [1.72275877725368... | Li4Al4 | cubic | 6.279846 | 146.014891 | 37.664442 | -21.680919 | 8 | Li4Al4_cubic |
| 18 | 1393 | LiAl_yace | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.044553 | 16.541594 | 87.130427 | -3.478909 | 1 | Al_fcc |
| 19 | 1406 | LiAl_yace | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | bcc | 3.953036 | 16.811334 | 72.667242 | -3.388831 | 1 | Al_bcc |
| 20 | 1419 | LiAl_yace | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | bcc | 4.216389 | 20.403222 | 15.823747 | -1.756104 | 1 | Li_bcc |
| 21 | 1435 | LiAl_yace | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | fcc | 4.331457 | 20.318983 | 14.231625 | -1.755594 | 1 | Li_fcc |
| 22 | 1451 | LiAl_yace | (Atom('Li', [4.5021943685456485, 2.599343130623782, 1.8380131542949232], index=0), Atom('Li', [6.753291552821257, 3.8990146959337566, 2.7570197314419675], index=1), Atom('Al', [-3.838851410290508e... | Li2Al2 | cubic | 6.367064 | 64.521799 | 46.107162 | -11.185880 | 4 | Li2Al2_cubic |
| 23 | 1464 | LiAl_yace | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [2.0106543994993293, 2.0106543994993293, 2.462341474538397e-16], index=1), Atom('Al', [2.0106543994993293, 1.2311707372691985e-16, 2.0106543994993... | LiAl3 | cubic | 5.686989 | 65.028366 | 66.254925 | -12.569153 | 4 | LiAl3_cubic |
| 24 | 1480 | LiAl_yace | (Atom('Li', [5.141009159558869, 1.0571139195527752, 0.820249453790277], index=0), Atom('Li', [3.2705789348169056, 1.5045550288016276, 2.715159327393234], index=1), Atom('Li', [-3.601125467999465, ... | Li9Al4 | monoclinic | 13.519944 | 213.136118 | 33.963240 | -31.796316 | 13 | Li9Al4_monoclinic |
| 25 | 1493 | LiAl_yace | (Atom('Al', [2.2270976540671734, 1.2858164055924044, 1.9025646270076813], index=0), Atom('Al', [-2.227095628822777, 3.8574462424884515, 2.7757665665986657], index=1), Atom('Li', [8.407589514518869... | Li3Al2 | trigonal | 6.299181 | 80.375104 | 39.643133 | -13.138303 | 5 | Li3Al2_trigonal |
| 26 | 1506 | LiAl_yace | (Atom('Li', [2.2269869888586107, 1.285751535686306, 7.864026721150146], index=0), Atom('Li', [-1.5554058443124377e-09, 2.571503074062492, 0.7130584901440213], index=1), Atom('Li', [-1.555405844312... | Li4Al4 | cubic | 6.298870 | 147.356944 | 46.701117 | -21.607231 | 8 | Li4Al4_cubic |
df_dft_ref = pd.read_pickle("dft_ref.pckl")
al_fcc = df_dft_ref[df_dft_ref["compound"]=="Al_fcc"]
li = df_dft_ref[df_dft_ref["compound"].isin(["Li_bcc","Li_fcc"])]
df_mixed = df_dft_ref[df_dft_ref["compound"].isin(["LiAl_mp-1067","LiAl3_mp-10890","Li9Al4_mp-568404","Li3Al2_mp-16506","LiAl_mp-1079240"])]
li["energy_per_atom"] = li["energy"]/li["number_of_atoms"]
# li
fig, ax_list = plt.subplots(ncols=3, nrows=len(potentials_list), sharex="col")
fig.set_figwidth(24)
fig.set_figheight(20)
color_palette = sns.color_palette("tab10", n_colors=len(data_murn.phase.unique()))
for i, pot in enumerate(potentials_list):
mask1 = data_murn["compound"]=="Al"
data1 = data_murn[(data_murn.potential == get_clean_project_name(pot)) & (mask1)]
mask2 = data_murn["compound"]=="Li"
data2 = data_murn[(data_murn.potential == get_clean_project_name(pot)) & (mask2)]
mask3 = data_murn["compound"].isin(["Al","Li"])
data3 = data_murn[(data_murn.potential == get_clean_project_name(pot)) & (~mask3)]
for j,(_, row) in enumerate(data1.iterrows()):
murn_job = pr.load(row["job_id"])
murn_df = murn_job.output_to_pandas()
n_atoms = row["n_atoms"]
ax_list[i,0].plot(murn_df["volume"]/n_atoms, murn_df["energy"]/n_atoms,"-",
lw=4,
label= row["phase"],
color=color_palette[j])
ax_list[i,0].set_title(f"{get_clean_project_name(pot)}" + '_' + data1.iloc[0]["compound"],fontsize=22)
ax_list[i,0].legend(prop={"size":16})
ax_list[i,0].scatter(al_fcc["vol"],al_fcc["energy"]/al_fcc["number_of_atoms"],
facecolor="none",edgecolor="k",s=100,label="DFT")
for j,(_, row) in enumerate(data2.iterrows()):
murn_job = pr.load(row["job_id"])
murn_df = murn_job.output_to_pandas()
n_atoms = row["n_atoms"]
ax_list[i,2].plot(murn_df["volume"]/n_atoms, murn_df["energy"]/n_atoms,"-",
lw=4,
label= row["phase"],
color=color_palette[j])
ax_list[i,2].set_title(f"{get_clean_project_name(pot)}" + '_' + data2.iloc[0]["compound"],fontsize=22)
# ax_list[i,2].legend(prop={"size":16})
ax_list[i,2].scatter(li["vol"],li["energy"]/li["number_of_atoms"],
facecolor="none",edgecolor="k",s=100,label="DFT")
for j,(_, row) in enumerate(data3.iterrows()):
murn_job = pr.load(row["job_id"])
murn_df = murn_job.output_to_pandas()
n_atoms = row["n_atoms"]
ax_list[i,1].plot(murn_df["volume"]/n_atoms, murn_df["energy"]/n_atoms,"-",
lw=4,
label= row["phase"],
color=color_palette[j])
ax_list[i,1].set_title(f"{get_clean_project_name(pot)}" + '_AlLi_mixed',fontsize=22)
# ax_list[i,1].legend(prop={"size":16})
ax_list[i,1].scatter(df_mixed["vol"],df_mixed["energy"]/df_mixed["number_of_atoms"],
facecolor="none",edgecolor="k",s=100,label="DFT")
for i in range(3):
ax_list[0,i].legend(prop={"size":16})
ax_list[-1,i].set_xlabel("Volume per atom, $\mathrm{\AA^3}$",fontsize=20)
ax_list[-1,i].tick_params(axis="x",labelsize=18)
for i in range(len(potentials_list)):
ax_list[i,0].set_ylabel("Energy per atom, eV/atom",fontsize=18)
# ax.legend(prop={"size":16})
# ax.set_ylabel("Energy per atom, eV/atom",fontsize=20)
#break
fig.subplots_adjust(wspace=0.1);
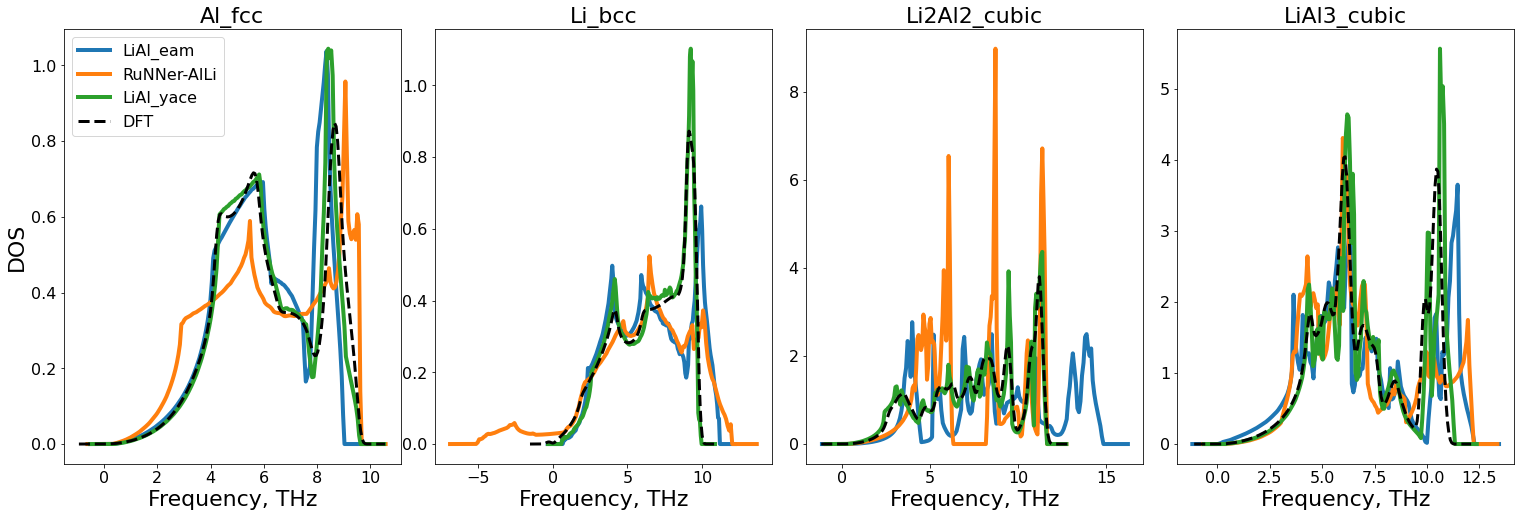
(b) Elastic constants and Phonons#
Pyiron also has job modules to calculate elastic constants and thermal properties using the quasi-harmonic approximation given by the phonopy package.
As in the previous task, we again loop over the defined potentials and then over the given structures.
Calculating elastic constants and thermal properties is considerably more expensive than calculating EV curves. Hence, it is useful to only calculate these properties for a subset of most important structures
list_of_phases = ["Al_fcc","Li_bcc","Li2Al2_cubic","LiAl3_cubic"]
subset_murn = data_murn[data_murn["phase"].isin(list_of_phases)]
subset_murn
| job_id | potential | ase_atoms | compound | crystal_structure | a | eq_vol | eq_bm | eq_energy | n_atoms | phase | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1140 | LiAl_eam | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.039967 | 16.495612 | 85.876912 | -3.483097 | 1 | Al_fcc |
| 2 | 1166 | LiAl_eam | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | bcc | 4.195477 | 20.114514 | 13.690609 | -1.757011 | 1 | Li_bcc |
| 4 | 1192 | LiAl_eam | (Atom('Li', [4.359978178265943, 2.5172345748814795, 1.7799536377360747], index=0), Atom('Li', [6.53996726740165, 3.775851862320358, 2.669930456604317], index=1), Atom('Al', [-3.964456982410852e-12... | Li2Al2 | cubic | 6.165940 | 58.604895 | 100.347240 | -11.074362 | 4 | Li2Al2_cubic |
| 5 | 1205 | LiAl_eam | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [1.9825515172760235, 1.9825515172760237, 2.427925369776811e-16], index=1), Atom('Al', [1.9825515172760235, 1.2139626848884054e-16, 1.9825515172760... | LiAl3 | cubic | 5.607502 | 62.227580 | 51.472656 | -12.774590 | 4 | LiAl3_cubic |
| 9 | 1257 | RuNNer-AlLi | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.025259 | 16.355737 | 76.669339 | -3.484016 | 1 | Al_fcc |
| 11 | 1283 | RuNNer-AlLi | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | bcc | 4.211118 | 20.286595 | 8.517306 | -1.755918 | 1 | Li_bcc |
| 13 | 1309 | RuNNer-AlLi | (Atom('Li', [4.509081801264686, 2.603319591757272, 1.8408249369278522], index=0), Atom('Li', [6.763622701898693, 3.90497938763465, 2.7612374053913604], index=1), Atom('Al', [-3.844724064520768e-12... | Li2Al2 | cubic | 6.376805 | 64.816143 | 57.934650 | -11.212634 | 4 | Li2Al2_cubic |
| 14 | 1322 | RuNNer-AlLi | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [2.0154153406879987, 2.0154153406879987, 2.46817194592603e-16], index=1), Atom('Al', [2.0154153406879987, 1.234085972963015e-16, 2.015415340687998... | LiAl3 | cubic | 5.700455 | 65.403086 | 59.308440 | -12.574696 | 4 | LiAl3_cubic |
| 18 | 1393 | LiAl_yace | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.044553 | 16.541594 | 87.130427 | -3.478909 | 1 | Al_fcc |
| 20 | 1419 | LiAl_yace | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | bcc | 4.216389 | 20.403222 | 15.823747 | -1.756104 | 1 | Li_bcc |
| 22 | 1451 | LiAl_yace | (Atom('Li', [4.5021943685456485, 2.599343130623782, 1.8380131542949232], index=0), Atom('Li', [6.753291552821257, 3.8990146959337566, 2.7570197314419675], index=1), Atom('Al', [-3.838851410290508e... | Li2Al2 | cubic | 6.367064 | 64.521799 | 46.107162 | -11.185880 | 4 | Li2Al2_cubic |
| 23 | 1464 | LiAl_yace | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [2.0106543994993293, 2.0106543994993293, 2.462341474538397e-16], index=1), Atom('Al', [2.0106543994993293, 1.2311707372691985e-16, 2.0106543994993... | LiAl3 | cubic | 5.686989 | 65.028366 | 66.254925 | -12.569153 | 4 | LiAl3_cubic |
for pot in potentials_list:
group_name = get_clean_project_name(pot)
pr_pot = pr.create_group(group_name)
print(group_name)
for _, row in subset_murn[subset_murn.potential==group_name].iterrows():
job_id = row["job_id"]
job_ref = pr_pot.create_job(pr_pot.job_type.Lammps, f"ref_job_{row.compound}_{row.crystal_structure}")
ref = pr_pot.load(job_id)
job_ref.structure = ref.structure
job_ref.potential = pot
job_ref.calc_minimize()
elastic_job = job_ref.create_job(pr_pot.job_type.ElasticMatrixJob, f"elastic_job_{row.compound}_{row.crystal_structure}")
elastic_job.input["eps_range"] = 0.05
elastic_job.run()
phonopy_job = job_ref.create_job(pr_pot.job_type.PhonopyJob, f"phonopy_job_{row.compound}_{row.crystal_structure}")
job_ref.calc_static()
phonopy_job.run()
LiAl_eam
2022-06-08 14:17:48,273 - pyiron_log - WARNING - The job elastic_job_Al_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:48,454 - pyiron_log - WARNING - The job phonopy_job_Al_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:48,754 - pyiron_log - WARNING - The job elastic_job_Li_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:48,922 - pyiron_log - WARNING - The job phonopy_job_Li_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:49,223 - pyiron_log - WARNING - The job elastic_job_Li2Al2_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:49,398 - pyiron_log - WARNING - The job phonopy_job_Li2Al2_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:49,706 - pyiron_log - WARNING - The job elastic_job_LiAl3_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:49,875 - pyiron_log - WARNING - The job phonopy_job_LiAl3_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
RuNNer-AlLi
2022-06-08 14:17:50,177 - pyiron_log - WARNING - The job elastic_job_Al_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:50,342 - pyiron_log - WARNING - The job phonopy_job_Al_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:50,639 - pyiron_log - WARNING - The job elastic_job_Li_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:50,802 - pyiron_log - WARNING - The job phonopy_job_Li_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:51,099 - pyiron_log - WARNING - The job elastic_job_Li2Al2_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:51,271 - pyiron_log - WARNING - The job phonopy_job_Li2Al2_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:51,581 - pyiron_log - WARNING - The job elastic_job_LiAl3_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:51,752 - pyiron_log - WARNING - The job phonopy_job_LiAl3_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
LiAl_yace
2022-06-08 14:17:52,054 - pyiron_log - WARNING - The job elastic_job_Al_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:52,218 - pyiron_log - WARNING - The job phonopy_job_Al_fcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:52,515 - pyiron_log - WARNING - The job elastic_job_Li_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:52,680 - pyiron_log - WARNING - The job phonopy_job_Li_bcc is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:52,983 - pyiron_log - WARNING - The job elastic_job_Li2Al2_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:53,159 - pyiron_log - WARNING - The job phonopy_job_Li2Al2_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:53,468 - pyiron_log - WARNING - The job elastic_job_LiAl3_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
2022-06-08 14:17:53,638 - pyiron_log - WARNING - The job phonopy_job_LiAl3_cubic is being loaded instead of running. To re-run use the argument 'delete_existing_job=True in create_job'
def filter_elastic(job_table):
return (job_table.hamilton == "ElasticMatrixJob") & (job_table.status == "finished")
# Get corresponding elastic constants
def get_c11(job_path):
return job_path["output/elasticmatrix"]["C"][0, 0]
def get_c12(job_path):
return job_path["output/elasticmatrix"]["C"][0, 1]
def get_c44(job_path):
return job_path["output/elasticmatrix"]["C"][3, 3]
table = pr.create_table("table_elastic", delete_existing_job=True)
table.db_filter_function = filter_elastic
table.add["potential"] = get_potential
table.add["C11"] = get_c11
table.add["C12"] = get_c12
table.add["C44"] = get_c44
table.add["compound"] = get_compound
table.add["crystal_structure"] = get_crystal_structure
table.run()
data_elastic = table.get_dataframe()
data_elastic["phase"] = data_elastic.compound + "_" + data_elastic.crystal_structure
data_elastic = data_elastic[data_elastic["phase"].isin(list_of_phases)]
data_elastic
The job table_elastic was saved and received the ID: 1780
| job_id | potential | C11 | C12 | C44 | compound | crystal_structure | phase | |
|---|---|---|---|---|---|---|---|---|
| 0 | 1524 | LiAl_eam | 120.339279 | 66.483631 | 45.515458 | Al | fcc | Al_fcc |
| 1 | 1540 | LiAl_eam | 16.740018 | 11.018163 | 12.688217 | Li | bcc | Li_bcc |
| 2 | 1556 | LiAl_eam | 179.464635 | 54.231219 | 47.889040 | Li2Al2 | cubic | Li2Al2_cubic |
| 3 | 1573 | LiAl_eam | 65.443987 | 47.601166 | 28.002138 | LiAl3 | cubic | LiAl3_cubic |
| 4 | 1590 | RuNNer-AlLi | 119.613688 | 59.261331 | 57.671025 | Al | fcc | Al_fcc |
| 5 | 1606 | RuNNer-AlLi | 13.974565 | 4.476591 | 13.293350 | Li | bcc | Li_bcc |
| 6 | 1622 | RuNNer-AlLi | 124.404880 | 20.665379 | 42.673693 | Li2Al2 | cubic | Li2Al2_cubic |
| 7 | 1639 | RuNNer-AlLi | 88.575923 | 50.190830 | 48.202184 | LiAl3 | cubic | LiAl3_cubic |
| 8 | 1685 | LiAl_yace | 133.807535 | 62.693651 | 40.423203 | Al | fcc | Al_fcc |
| 9 | 1701 | LiAl_yace | 18.307762 | 13.775557 | 12.106574 | Li | bcc | Li_bcc |
| 10 | 1717 | LiAl_yace | 114.275413 | 13.925574 | 42.537995 | Li2Al2 | cubic | Li2Al2_cubic |
| 11 | 1734 | LiAl_yace | 112.037951 | 42.770574 | 45.206508 | LiAl3 | cubic | LiAl3_cubic |
fig, ax_list = plt.subplots(ncols=len(data_elastic.phase.unique()), nrows=1,)
fig.set_figwidth(26)
fig.set_figheight(8)
color_palette = sns.color_palette("tab10", n_colors=len(data_elastic.potential.unique()))
pot = "LiAl_yace"
for i, phase in enumerate(data_elastic.phase.unique()):
ax = ax_list[i]
# data = data_elastic[(data_elastic.phase == phase) & (data_elastic["potential"]=="pot")]
data = data_elastic[(data_elastic.phase == phase)]
# DFT data is read from csv files
dft_ref = pd.read_csv(phase.lower()+"_dos.csv")
for j, pot in enumerate(potentials_list):
phonopy_job = pr[get_clean_project_name(pot) + f"/phonopy_job_{phase}"]
thermo = phonopy_job.get_thermal_properties(t_min=0, t_max=800)
ax.plot(phonopy_job["output/dos_energies"], phonopy_job["output/dos_total"],
lw=4,
color=color_palette[j],
label=get_clean_project_name(pot))
ax.set_xlabel("Frequency, THz",fontsize=22)
ax.plot(dft_ref["dos_energy"],dft_ref["dos_total"],ls="--",lw=3,color="k",label="DFT")
ax.set_title(f"{phase}",fontsize=22)
ax.tick_params(labelsize=16)
ax_list[0].set_ylabel("DOS",fontsize=22)
ax_list[0].legend(prop={"size":16})
fig.subplots_adjust(wspace=0.1);
# fig, ax_list = plt.subplots(ncols=len(data_elastic.phase.unique()), nrows=len(potentials_list), sharey="row")
# fig.set_figwidth(25)
# fig.set_figheight(12)
# color_palette = sns.color_palette("tab10", n_colors=len(data_elastic.potential.unique()))
# for i, phase in enumerate(data_elastic.phase.unique()):
# data = data_elastic[data_elastic.phase == phase]
# for j, pot in enumerate(potentials_list):
# ax = ax_list[j][i]
# phonopy_job = pr[get_clean_project_name(pot) + f"/phonopy_job_{phase}"]
# phonopy_job.plot_band_structure(axis=ax)
# ax.set_ylabel("")
# ax.set_title(get_clean_project_name(pot)+"__"+phase,fontsize=18)
# ax_list[j][0].set_ylabel("DOS")
# # ax_list[0][i].set_title(f"{phase}")
# fig.subplots_adjust(wspace=0.1, hspace=0.4);
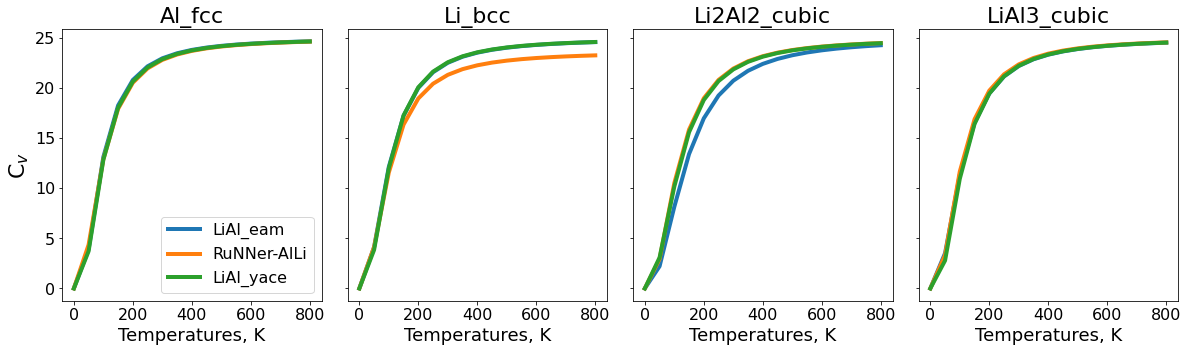
fig, ax_list = plt.subplots(ncols=len(data_elastic.phase.unique()), nrows=1, sharex="row", sharey="row")
fig.set_figwidth(20)
fig.set_figheight(5)
color_palette = sns.color_palette("tab10", n_colors=len(data_elastic.potential.unique()))
for i, phase in enumerate(data_elastic.phase.unique()):
ax = ax_list[i]
data = data_elastic[data_elastic.phase == phase]
n_atom = data_murn[data_murn["phase"]==phase]["n_atoms"].iloc[0]
for j, pot in enumerate(potentials_list):
phonopy_job = pr[get_clean_project_name(pot) + f"/phonopy_job_{phase}"]
thermo = phonopy_job.get_thermal_properties(t_min=0, t_max=800)
ax.plot(thermo.temperatures, thermo.cv/n_atom,
lw=4,
label=get_clean_project_name(pot),
color=color_palette[j])
ax.set_xlabel("Temperatures, K",fontsize=18)
ax.set_title(f"{phase}",fontsize=22)
ax.tick_params(labelsize=16)
ax_list[0].set_ylabel("C$_v$",fontsize=22)
ax_list[0].legend(prop={"size":16})
fig.subplots_adjust(wspace=0.1);
# phonopy_job.plot_band_structure()
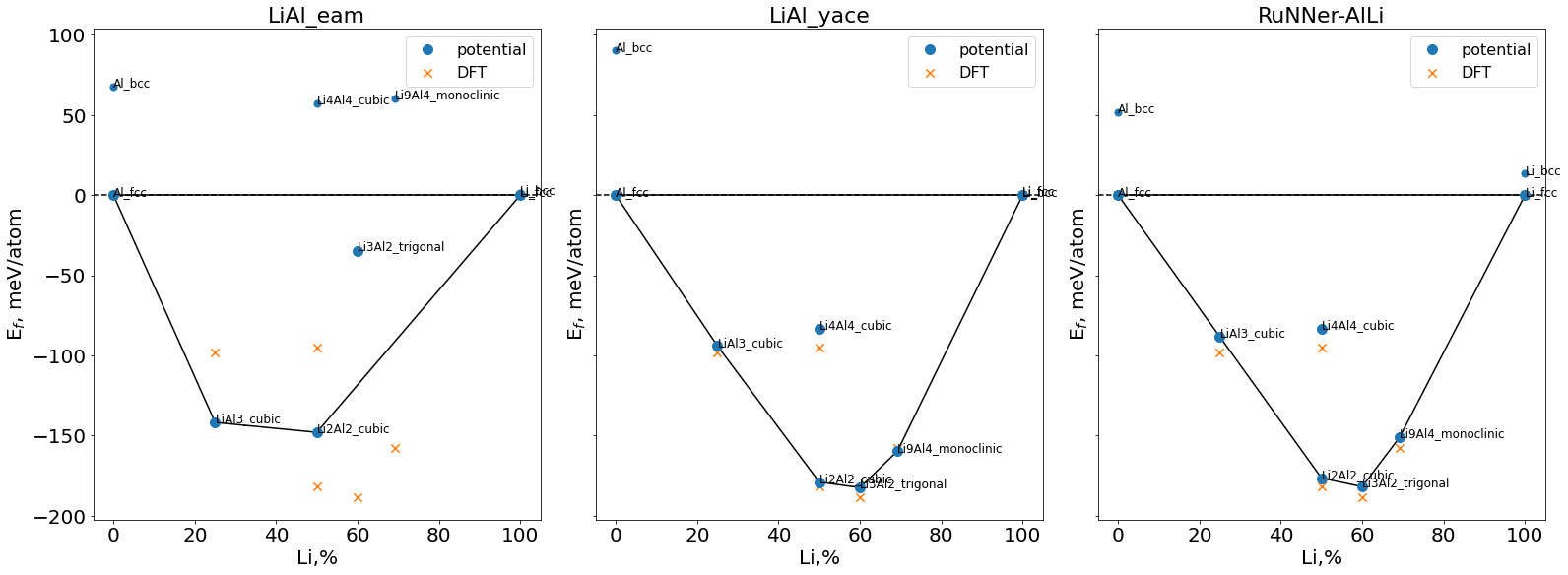
(c) Convex hull#
To assess the stability of the binary phases, we plot a convex hull for the considered phases.
For this task we compute the formation energies of the mixed phases relative to ground state energies of equilibrium unary phases.
from collections import Counter
# pot = "LiAl_yace"
# data_convexhull = data_murn[data_murn["potential"]==pot]
data_convexhull = data_murn.copy()
data_convexhull.head(2)
| job_id | potential | ase_atoms | compound | crystal_structure | a | eq_vol | eq_bm | eq_energy | n_atoms | phase | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1140 | LiAl_eam | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.039967 | 16.495612 | 85.876912 | -3.483097 | 1 | Al_fcc |
| 1 | 1153 | LiAl_eam | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | bcc | 3.898853 | 16.147864 | 48.620841 | -3.415312 | 1 | Al_bcc |
Using Collections.counter we construct a composition dictionary for all the phases and from that dictionary, we can extract the relative concentrations of Al and Li in each structure
Obtain the equilibrium energies for unary Al and Li phases from the Dataframe
Calculate the relative formation energies by subtracting the total energies of the mixed phases with the energies of eq Al and Li
Similarly calculate the formation energies from DFT ref data
def get_e_form(data_convexhull):
data_convexhull["comp_dict"] = data_convexhull["ase_atoms"].map(lambda at: Counter(at.get_chemical_symbols()))
data_convexhull["n_Al"] = data_convexhull["comp_dict"].map(lambda d: d.get("Al",0))
data_convexhull["n_Li"] = data_convexhull["comp_dict"].map(lambda d: d.get("Li",0))
data_convexhull["cAl"]= data_convexhull["n_Al"]/data_convexhull["n_atoms"] * 100
data_convexhull["cLi"]= data_convexhull["n_Li"]/data_convexhull["n_atoms"] * 100
E_f_Al = data_convexhull.loc[data_convexhull["n_Li"]==0,"eq_energy"].min()
E_f_Li = data_convexhull.loc[data_convexhull["n_Al"]==0,"eq_energy"].min()
data_convexhull["E_form"]=(data_convexhull["eq_energy"])-(data_convexhull[["n_Al","n_Li"]].values * [E_f_Al, E_f_Li]).sum(axis=1)
data_convexhull["E_form_per_atom"] = data_convexhull["E_form"]/data_convexhull["n_atoms"] * 1e3
data_convexhull = data_convexhull.sort_values("cLi")
return data_convexhull
df_eam = get_e_form(data_murn[data_murn["potential"]=="LiAl_eam"].copy())
df_nnp = get_e_form(data_murn[data_murn["potential"]=="RuNNer-AlLi"].copy())
df_ace = get_e_form(data_murn[data_murn["potential"]=="LiAl_yace"].copy())
data_convexhull = pd.concat([df_eam,df_nnp,df_ace])
data_convexhull
| job_id | potential | ase_atoms | compound | crystal_structure | a | eq_vol | eq_bm | eq_energy | n_atoms | phase | comp_dict | n_Al | n_Li | cAl | cLi | E_form | E_form_per_atom | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1140 | LiAl_eam | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.039967 | 16.495612 | 85.876912 | -3.483097 | 1 | Al_fcc | {'Al': 1} | 1 | 0 | 100.000000 | 0.000000 | 0.000000 | 0.000000 |
| 1 | 1153 | LiAl_eam | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | bcc | 3.898853 | 16.147864 | 48.620841 | -3.415312 | 1 | Al_bcc | {'Al': 1} | 1 | 0 | 100.000000 | 0.000000 | 0.067785 | 67.785186 |
| 5 | 1205 | LiAl_eam | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [1.9825515172760235, 1.9825515172760237, 2.427925369776811e-16], index=1), Atom('Al', [1.9825515172760235, 1.2139626848884054e-16, 1.9825515172760... | LiAl3 | cubic | 5.607502 | 62.227580 | 51.472656 | -12.774590 | 4 | LiAl3_cubic | {'Li': 1, 'Al': 3} | 3 | 1 | 75.000000 | 25.000000 | -0.567192 | -141.797976 |
| 4 | 1192 | LiAl_eam | (Atom('Li', [4.359978178265943, 2.5172345748814795, 1.7799536377360747], index=0), Atom('Li', [6.53996726740165, 3.775851862320358, 2.669930456604317], index=1), Atom('Al', [-3.964456982410852e-12... | Li2Al2 | cubic | 6.165940 | 58.604895 | 100.347240 | -11.074362 | 4 | Li2Al2_cubic | {'Li': 2, 'Al': 2} | 2 | 2 | 50.000000 | 50.000000 | -0.591954 | -147.988453 |
| 8 | 1244 | LiAl_eam | (Atom('Li', [2.142967147985671, 1.2372426587287435, 7.662120717536293], index=0), Atom('Li', [-8.783761113500244e-10, 2.4744853189563414, 0.5913679335098909], index=1), Atom('Li', [-8.783761113500... | Li4Al4 | cubic | 6.061226 | 131.389799 | 71.221355 | -20.506570 | 8 | Li4Al4_cubic | {'Li': 4, 'Al': 4} | 4 | 4 | 50.000000 | 50.000000 | 0.458247 | 57.280860 |
| 7 | 1231 | LiAl_eam | (Atom('Al', [2.1548001975659234, 1.244075358781918, 1.861784175000869], index=0), Atom('Al', [-2.154798282819334, 3.732223313213554, 2.6646760238080542], index=1), Atom('Li', [8.560563403365654e-0... | Li3Al2 | trigonal | 6.094693 | 72.810229 | 69.231669 | -12.413856 | 5 | Li3Al2_trigonal | {'Al': 2, 'Li': 3} | 2 | 3 | 40.000000 | 60.000000 | -0.173341 | -34.668107 |
| 6 | 1218 | LiAl_eam | (Atom('Li', [4.9874611628416465, 1.0099045365192156, 0.8188840806477526], index=0), Atom('Li', [3.1237816780987666, 1.455730745331952, 2.673723152073369], index=1), Atom('Li', [-3.4421956688209843... | Li9Al4 | monoclinic | 13.023701 | 190.504374 | 53.125276 | -28.970054 | 13 | Li9Al4_monoclinic | {'Li': 9, 'Al': 4} | 4 | 9 | 30.769231 | 69.230769 | 0.785300 | 60.407664 |
| 2 | 1166 | LiAl_eam | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | bcc | 4.195477 | 20.114514 | 13.690609 | -1.757011 | 1 | Li_bcc | {'Li': 1} | 0 | 1 | 0.000000 | 100.000000 | 0.001096 | 1.096047 |
| 3 | 1179 | LiAl_eam | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | fcc | 4.253841 | 19.241330 | 13.985972 | -1.758107 | 1 | Li_fcc | {'Li': 1} | 0 | 1 | 0.000000 | 100.000000 | 0.000000 | 0.000000 |
| 9 | 1257 | RuNNer-AlLi | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.025259 | 16.355737 | 76.669339 | -3.484016 | 1 | Al_fcc | {'Al': 1} | 1 | 0 | 100.000000 | 0.000000 | 0.000000 | 0.000000 |
| 10 | 1270 | RuNNer-AlLi | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | bcc | 3.958447 | 16.870137 | 51.052272 | -3.432183 | 1 | Al_bcc | {'Al': 1} | 1 | 0 | 100.000000 | 0.000000 | 0.051832 | 51.832389 |
| 14 | 1322 | RuNNer-AlLi | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [2.0154153406879987, 2.0154153406879987, 2.46817194592603e-16], index=1), Atom('Al', [2.0154153406879987, 1.234085972963015e-16, 2.015415340687998... | LiAl3 | cubic | 5.700455 | 65.403086 | 59.308440 | -12.574696 | 4 | LiAl3_cubic | {'Li': 1, 'Al': 3} | 3 | 1 | 75.000000 | 25.000000 | -0.353389 | -88.347230 |
| 13 | 1309 | RuNNer-AlLi | (Atom('Li', [4.509081801264686, 2.603319591757272, 1.8408249369278522], index=0), Atom('Li', [6.763622701898693, 3.90497938763465, 2.7612374053913604], index=1), Atom('Al', [-3.844724064520768e-12... | Li2Al2 | cubic | 6.376805 | 64.816143 | 57.934650 | -11.212634 | 4 | Li2Al2_cubic | {'Li': 2, 'Al': 2} | 2 | 2 | 50.000000 | 50.000000 | -0.706083 | -176.520795 |
| 17 | 1361 | RuNNer-AlLi | (Atom('Li', [2.220260976080854, 1.2818682724036983, 7.872085429446316], index=0), Atom('Li', [1.722758777253687e-10, 2.5637365444716322, 0.6790950189344616], index=1), Atom('Li', [1.72275877725368... | Li4Al4 | cubic | 6.279846 | 146.014891 | 37.664442 | -21.680919 | 8 | Li4Al4_cubic | {'Li': 4, 'Al': 4} | 4 | 4 | 50.000000 | 50.000000 | -0.667816 | -83.477017 |
| 16 | 1348 | RuNNer-AlLi | (Atom('Al', [2.2338755345732753, 1.289729472183878, 1.9126243306628208], index=0), Atom('Al', [-2.233873547699001, 3.869185551846968, 2.7799443936883206], index=1), Atom('Li', [9.007133262260959e-... | Li3Al2 | trigonal | 6.318351 | 81.143544 | 44.574696 | -13.185198 | 5 | Li3Al2_trigonal | {'Al': 2, 'Li': 3} | 2 | 3 | 40.000000 | 60.000000 | -0.909387 | -181.877324 |
| 15 | 1335 | RuNNer-AlLi | (Atom('Li', [5.206051477294367, 1.0619663179427192, 0.8311820920214751], index=0), Atom('Li', [3.28638171437237, 1.5211864250363467, 2.7226207058417775], index=1), Atom('Li', [-3.6198784902055765,... | Li9Al4 | monoclinic | 13.640614 | 218.932018 | 33.874957 | -31.820765 | 13 | Li9Al4_monoclinic | {'Li': 9, 'Al': 4} | 4 | 9 | 30.769231 | 69.230769 | -1.961363 | -150.874092 |
| 11 | 1283 | RuNNer-AlLi | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | bcc | 4.211118 | 20.286595 | 8.517306 | -1.755918 | 1 | Li_bcc | {'Li': 1} | 0 | 1 | 0.000000 | 100.000000 | 0.013342 | 13.341610 |
| 12 | 1296 | RuNNer-AlLi | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | fcc | 3.967043 | 15.678901 | 147.215464 | -1.769260 | 1 | Li_fcc | {'Li': 1} | 0 | 1 | 0.000000 | 100.000000 | 0.000000 | 0.000000 |
| 18 | 1393 | LiAl_yace | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | fcc | 4.044553 | 16.541594 | 87.130427 | -3.478909 | 1 | Al_fcc | {'Al': 1} | 1 | 0 | 100.000000 | 0.000000 | 0.000000 | 0.000000 |
| 19 | 1406 | LiAl_yace | (Atom('Al', [0.0, 0.0, 0.0], index=0)) | Al | bcc | 3.953036 | 16.811334 | 72.667242 | -3.388831 | 1 | Al_bcc | {'Al': 1} | 1 | 0 | 100.000000 | 0.000000 | 0.090078 | 90.077889 |
| 23 | 1464 | LiAl_yace | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [2.0106543994993293, 2.0106543994993293, 2.462341474538397e-16], index=1), Atom('Al', [2.0106543994993293, 1.2311707372691985e-16, 2.0106543994993... | LiAl3 | cubic | 5.686989 | 65.028366 | 66.254925 | -12.569153 | 4 | LiAl3_cubic | {'Li': 1, 'Al': 3} | 3 | 1 | 75.000000 | 25.000000 | -0.376321 | -94.080320 |
| 22 | 1451 | LiAl_yace | (Atom('Li', [4.5021943685456485, 2.599343130623782, 1.8380131542949232], index=0), Atom('Li', [6.753291552821257, 3.8990146959337566, 2.7570197314419675], index=1), Atom('Al', [-3.838851410290508e... | Li2Al2 | cubic | 6.367064 | 64.521799 | 46.107162 | -11.185880 | 4 | Li2Al2_cubic | {'Li': 2, 'Al': 2} | 2 | 2 | 50.000000 | 50.000000 | -0.715855 | -178.963696 |
| 26 | 1506 | LiAl_yace | (Atom('Li', [2.2269869888586107, 1.285751535686306, 7.864026721150146], index=0), Atom('Li', [-1.5554058443124377e-09, 2.571503074062492, 0.7130584901440213], index=1), Atom('Li', [-1.555405844312... | Li4Al4 | cubic | 6.298870 | 147.356944 | 46.701117 | -21.607231 | 8 | Li4Al4_cubic | {'Li': 4, 'Al': 4} | 4 | 4 | 50.000000 | 50.000000 | -0.667180 | -83.397512 |
| 25 | 1493 | LiAl_yace | (Atom('Al', [2.2270976540671734, 1.2858164055924044, 1.9025646270076813], index=0), Atom('Al', [-2.227095628822777, 3.8574462424884515, 2.7757665665986657], index=1), Atom('Li', [8.407589514518869... | Li3Al2 | trigonal | 6.299181 | 80.375104 | 39.643133 | -13.138303 | 5 | Li3Al2_trigonal | {'Al': 2, 'Li': 3} | 2 | 3 | 40.000000 | 60.000000 | -0.912174 | -182.434806 |
| 24 | 1480 | LiAl_yace | (Atom('Li', [5.141009159558869, 1.0571139195527752, 0.820249453790277], index=0), Atom('Li', [3.2705789348169056, 1.5045550288016276, 2.715159327393234], index=1), Atom('Li', [-3.601125467999465, ... | Li9Al4 | monoclinic | 13.519944 | 213.136118 | 33.963240 | -31.796316 | 13 | Li9Al4_monoclinic | {'Li': 9, 'Al': 4} | 4 | 9 | 30.769231 | 69.230769 | -2.075747 | -159.672835 |
| 20 | 1419 | LiAl_yace | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | bcc | 4.216389 | 20.403222 | 15.823747 | -1.756104 | 1 | Li_bcc | {'Li': 1} | 0 | 1 | 0.000000 | 100.000000 | 0.000000 | 0.000000 |
| 21 | 1435 | LiAl_yace | (Atom('Li', [0.0, 0.0, 0.0], index=0)) | Li | fcc | 4.331457 | 20.318983 | 14.231625 | -1.755594 | 1 | Li_fcc | {'Li': 1} | 0 | 1 | 0.000000 | 100.000000 | 0.000509 | 0.509341 |
Read df which contains DFT ref data for plotting
convex_ref = pd.read_pickle("dft_convexhull_ref.pckl")
convex_ref
| name | energy | vol | compound | ao | number_of_atoms | comp_dict | n_Al | n_Li | cAl | cLi | E_form | E_form_per_atom | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 438 | /home/users/lysogy36/tools/VASP/Al-Li/DFT/Al_fcc/murn/strain_1_0/data.json | -13.930995 | 16.484415 | Al_fcc | (Atom('Al', [0.0, 0.0, 0.0], index=0), Atom('Al', [0.0, 2.019983601551115, 2.019983601551115], index=1), Atom('Al', [2.019983601551115, 0.0, 2.019983601551115], index=2), Atom('Al', [2.01998360155... | 4 | {'Al': 4} | 4 | 0 | 100.000000 | 0.000000 | 0.000000 | 0.000000 |
| 910 | /home/users/lysogy36/tools/VASP/Al-Li/DFT/LiAl3_mp-10890/murn/strain_1_0/data.json | -12.597018 | 16.295840 | LiAl3_mp-10890 | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Al', [2.0122514573524146, 2.0122514573524146, 0.0], index=1), Atom('Al', [2.0122514573524146, 0.0, 2.0122514573524146], index=2), Atom('Al', [0.0, 2.01... | 4 | {'Li': 1, 'Al': 3} | 3 | 1 | 75.000000 | 25.000000 | -0.392474 | -98.118408 |
| 1950 | /home/users/lysogy36/tools/VASP/Al-Li/DFT/LiAl_mp-1067/murn/strain_1_0/data.json | -11.204795 | 16.028228 | LiAl_mp-1067 | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Li', [2.246243529971499, 1.2968693066945, 0.9170250810763773], index=1), Atom('Al', [4.492487059942998, 2.593738613389, 1.8340501621527545], index=2), ... | 4 | {'Li': 2, 'Al': 2} | 2 | 2 | 50.000000 | 50.000000 | -0.726701 | -181.675339 |
| 1275 | /home/users/lysogy36/tools/VASP/Al-Li/DFT/LiAl_mp-1079240/murn/strain_1_0/data.json | -21.715330 | 18.537039 | LiAl_mp-1079240 | (Atom('Li', [-2.093764484173552e-06, 2.574581270471953, 3.588630766943668], index=0), Atom('Li', [2.229653899294873, 1.2872887040708958, 5.022609593138096], index=1), Atom('Li', [2.229653899294873... | 8 | {'Li': 4, 'Al': 4} | 4 | 4 | 50.000000 | 50.000000 | -0.759143 | -94.892853 |
| 652 | /home/users/lysogy36/tools/VASP/Al-Li/DFT/Li3Al2_mp-16506/murn/strain_1_0/data.json | -13.176984 | 16.098544 | Li3Al2_mp-16506 | (Atom('Li', [7.387307289355338, 3.3557842846492325, 2.205190367378745], index=0), Atom('Li', [4.984874333407062, 2.2644466100798333, 1.488038392346624], index=1), Atom('Li', [0.0, 0.0, 0.0], index... | 5 | {'Li': 3, 'Al': 2} | 2 | 3 | 40.000000 | 60.000000 | -0.942593 | -188.518538 |
| 231 | /home/users/lysogy36/tools/VASP/Al-Li/DFT/Li9Al4_mp-568404/murn/strain_1_0/data.json | -31.786765 | 16.532577 | Li9Al4_mp-568404 | (Atom('Li', [15.085585487572331, 3.6087478779487228, 4.372653838370371], index=0), Atom('Li', [13.209884188064274, 3.160045831227256, 2.4668794892606694], index=1), Atom('Li', [6.31626414433567, 1... | 13 | {'Li': 9, 'Al': 4} | 4 | 9 | 30.769231 | 69.230769 | -2.049089 | -157.622253 |
| 1343 | /home/users/lysogy36/tools/VASP/Al-Li/DFT/Li_bcc/murn/strain_1_0/data.json | -3.512596 | 20.099126 | Li_bcc | (Atom('Li', [0.0, 0.0, 0.0], index=0), Atom('Li', [1.712796338409787, 1.712796338409787, 1.712796338409787], index=1)) | 2 | {'Li': 2} | 0 | 2 | 0.000000 | 100.000000 | 0.000000 | 0.000000 |
Define a function to automatically get the mathematical convex hull
from scipy.spatial import ConvexHull,convex_hull_plot_2d
def get_convexhull(df):
df_tmp = df.reset_index()
points = np.zeros([len(df_tmp["cLi"]),2])
for i,row in df_tmp.iterrows():
points[i,0], points[i,1] = float(row["cLi"]), float(row["E_form_per_atom"])
hull = ConvexHull(points)
return hull,points
fig,ax = plt.subplots(figsize=(22,8),ncols=len(potentials_list),constrained_layout=True,sharey="row")
dfs = ([pd.DataFrame(y) for x, y in data_convexhull.groupby(by='potential', as_index=False)])
for i,pot in enumerate(potentials_list):
df_tmp = dfs[i].copy()
ax[i].scatter(df_tmp["cLi"],df_tmp["E_form_per_atom"],marker="o",s=50)
df_tmp = df_tmp[(df_tmp["E_form_per_atom"]<0.1) & (df_tmp["E_form"]<0.1)]
hull,points = get_convexhull(df_tmp)
for simplex in hull.simplices:
ax[i].plot(points[simplex, 0], points[simplex, 1], 'k-')
ax[i].axhline(0,ls="--",color="k")
ax[i].plot(df_tmp["cLi"], df_tmp["E_form_per_atom"],"o",markersize=10,label="potential")
ax[i].scatter(convex_ref["cLi"],convex_ref["E_form_per_atom"],marker="x",s=70,
label="DFT")
ax[i].legend(prop={"size":16})
ax[i].set_xlabel("Li,%",fontsize="20")
ax[i].set_ylabel("E$_f$, meV/atom",fontsize="20")
ax[i].tick_params(labelsize=20,axis="both")
# ax.set_ylim(-200,10)
for _,row in dfs[i].iterrows():
ax[i].text((row["cLi"]+0.01),row["E_form_per_atom"],row["phase"],size=12)
ax[i].set_title(dfs[i].iloc[0]["potential"],fontsize=22)
plt.show()
time_stop = time.time()
print(f"Total run time for the notebook {time_stop - time_start} seconds")
Total run time for the notebook 180.9642095565796 seconds

